by Shannon S.Zhang, Misako Nagasaka, Viola W.Zhu and Sai-Hong Ignatius Ou
This recent review from August, 2021 highlights what we know about EML4-ALK variants and TP53 mutations clinically. Authors are comprised of researchers in the US (Orange, CA and Detroit, MI) and Japan (Kawasaki, Kanagawa).
After the discovery in 2007 that anaplastic lymphoma kinase (ALK) was altered in non-small cell lung cancer, there have been 6 approved tyrosine kinase inhibitors approved globally. Usually, when a sample is diagnosed by fluorescence in situ hybridization (FISH) or immunohistochemistry (IHC), information such as fusion partner or fusion break location cannot be ascertained. By screening patients using next-generation sequencing (ngs), the fusion partner and definitive break points can be identified. Over 90 fusion partners are known for ALK in NSCLC. The majority (85%) are with EML4-ALK variants.
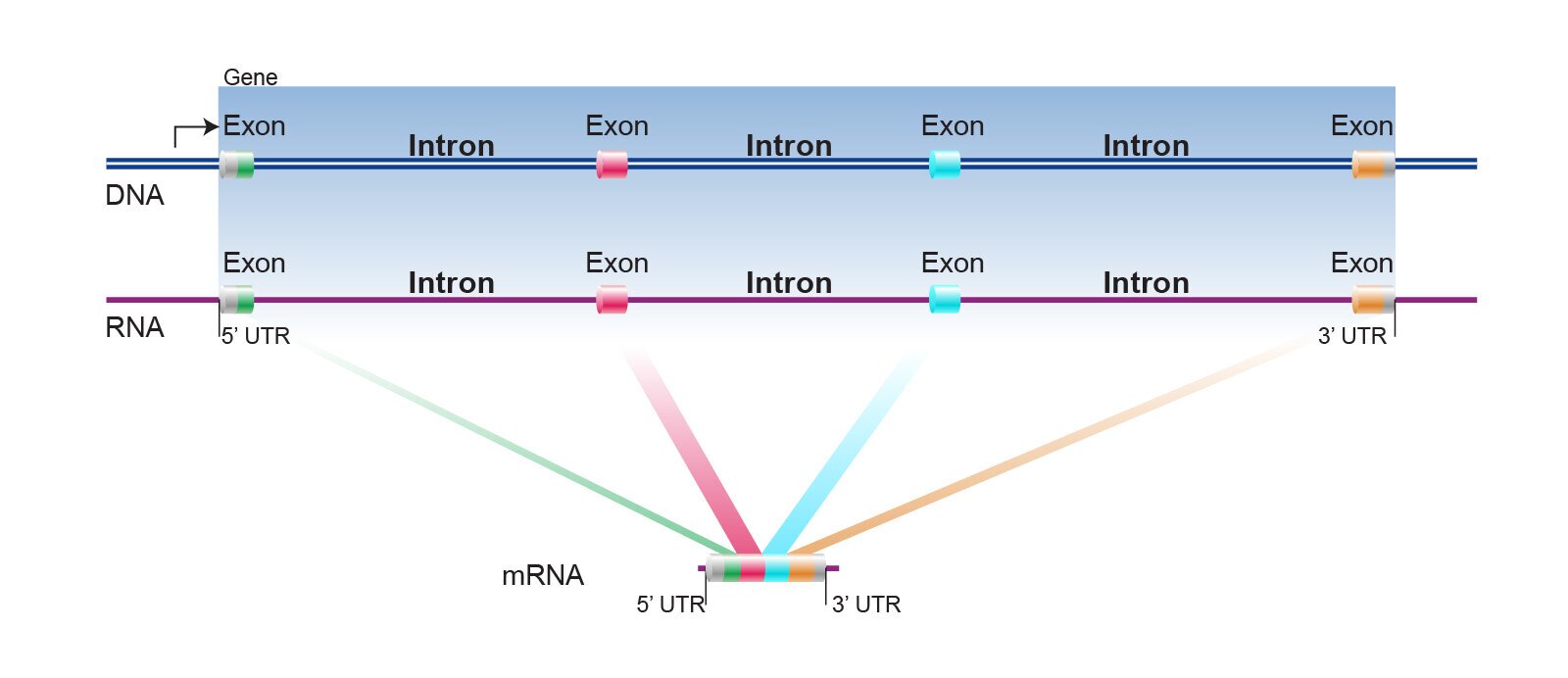
In a DNA sequence, not all coded amino acids confer to a protein.There are structures for regulation that are cut out before the RNA is translated into a protein.The “exons” are coded and “introns” are cut out. Just think of it like a train. The train sections are exons and the connectors or linkers are introns.
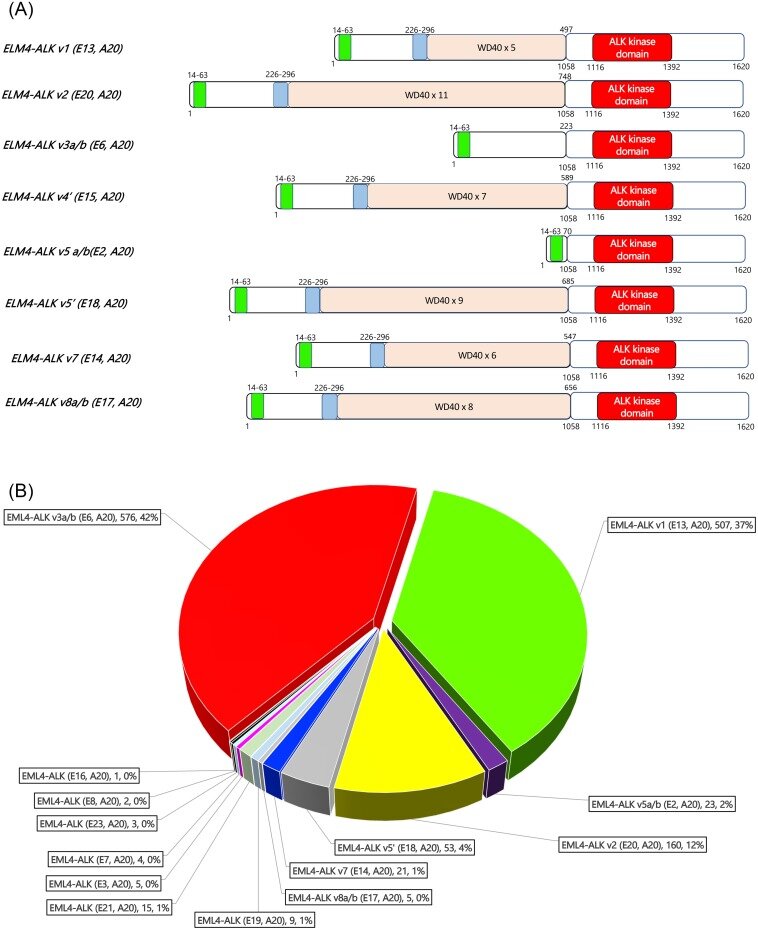
EML4 contains 26 exons and there are many different fusion partners with various deletions and insertions. The majority have been found to have exon 1-13 fused with exon 20 of ALK. This was the original version described in 2007 and it was called variant 1. Variant 2 and variant 3 were sequentially published, however at this point there are many variants. The majority tend to be variant 1, variant 3 and variant 5.
A “simple” explanation is to think of a shorter and a longer variant according to how long the resultant fusion becomes. Short versions tend to not have a “TAPE” domain from EML4, and the longer versions tend to retain the “TAPE” domain. This “TAPE” domain helps EML4 to interact with other proteins and is very hydrophobic in nature.
Variant 1 (EML4 exon13-joined with ALK exon20) and variant 3 (EML4 exon6 joined with ALK exon20) make up the majority of the EML4-ALK fusions that have been published.
In the laboratory, different variants will convey different stability of this fusion protein in the cell. The stability of the EML4-ALK fusion protein may determine how long a patient remains progression free under treatment with various TKIs. Through various published data, the authors reported (Japan, China, Taiwan, Germany, France, US and South Korea) that usually variant 1 patients tend to fair better than variant 3 patients. It is hypothesized that an ALK variant 3 patient may be more susceptible to an HSP90 inhibitor. The authors recommend lorlatinib as the first line treatment for variant 3 patients
ALK NSCLC patients very often have a concurrent TP53 mutation. The frequency of TP53 mutations is not affected by location, sex, age or stage of diagnosis (Germany, China, France, Japan, or US). Some researchers have reasoned that ALK mutation variant 3 plus TP53 mutation is a bad recipe. However, ALK patients are not just statistics because some patients with these combinations are still doing well.
https://www.sciencedirect.com/science/article/pii/S0169500221004396
https://www.genome.gov/genetics-glossary/Intron
Author: Alice Chou
September 12, 2021



